Molecular Docking of Quinine, Chloroquine and Hydroxychloroquine to Angiotensin Converting Enzyme 2 (ACE2) Receptor for Discovering New Potential COVID-19 Antidote
Keri Lestari1*, Trully Sitorus2, Instiaty3, Sandra Megantara4, Jutti Levita1
1Department of Pharmacology and Clinical Pharmacy, Universitas Padjadjaran, Indonesia. 2Division of Pharmacology and Therapy, Department of Biomedical Sciences, Universitas Padjadjaran, Indonesia. 3Department of Pharmacology and Therapeutics, Faculty of Medicine, Universitas Indonesia, Indonesia. 4Department of Pharmaceutical Analysis and Medicinal Chemistry, Universitas Padjadjaran, Indonesia.
Correspondence: Keri Lestari, Department of Pharmacology and Clinical Pharmacy, Universitas Padjadjaran, Indonesia. Email: lestarikd @ unpad.ac.id
|
ABSTRACT Researches on drugs to be used for COVID-19 therapy are still challenging. SARS-CoV-2 uses the angiotensin-converting enzyme (ACE2) receptor to enter the human body. The S (spike) protein structure of SARS-CoV-2 interacts with the active site of ACE2 receptor, defined as the peptidase domain, which consists of Gln24, Asp30, His34, Tyr41, Gln42, Met82, Lys353, Arg357. This work studied the interaction of three quinoline-based antimalarial drugs with the peptidase domain of ACE2 receptor. The X-ray crystal structure of human ACE2 receptor was downloaded from Protein Data Bank. The ligands were built using MarvinSketch and were geometry optimized using MMFF94 in LigandScout. The energy-minimized ligands were docked to the peptidase domain of ACE2 receptor. Results showed that chloroquine, hydroxychloroquine, and quinine can interact with amino acid residues in the peptidase domain of the ACE2 receptor. Of the three compounds, quinine shows the strongest affinity to the ACE2 receptor (-4.89 kcal/mol) followed by hydroxychloroquine (-3.87 kcal/mol), and chloroquine (-3.17 kcal/mol), respectively. In conclusion, quinine, chloroquine, and hydroxychloroquine could block the infection of the SARS-CoV-2 virus by interacting with residue Lys353 in the peptidase domain of ACE2 receptor, thus potential to be used as COVID-19 antidote. This study will add more insights to the mechanism of quinoline-based antimalarial drugs in inhibiting the infection of the SARS-CoV-2 virus. Keywords: ACE2 receptor, COVID-19, Chloroquine, Hydroxychloroquine, Quinine, SARS-CoV-2 |
Introduction
SARS-CoV-2, a novel virus that emerged from Wuhan, China, is responsible for an outbreak of respiratory illness. The disease
it causes is popularly known as coronavirus disease-19 or COVID-19. SARS-CoV-2 uses the angiotensin-converting enzyme (ACE2) to enter the human body [1]. The ACE2 receptor comprises of an N-terminal peptidase domain and a C-terminal collectrin-like domain that ends with a single transmembrane helix and a~40-residue intracellular segment. The S (spike) protein structure of SARS-CoV-2 interacts with the active site of ACE2 receptor (Kd of~15nM) [2]. The key amino acid residues in the active peptidase domain of ACE2 receptor (the site where SARS-CoV-2 attaches) are Gln24, Asp30, His34, Tyr41, Gln42, Met82, Lys353, Arg357 [3].
On February 17, 2020, the Chinese State Council confirmed that chloroquine phosphate, an old drug for the treatment of malaria, had demonstrated a tangible and safe activity in treating pneumonia related to COVID-19. The clinical trial was conducted in a multicentre clinic in China. The infected patients who were given chloroquine experienced a more rapid recovery in fever and an improvement of lung computed tomography images [4,5].
In a recent in vitro study, chloroquine was found to block COVID-19 infection at low micromolar concentrations, with a maximum effective concentration (EC50) of 1.13 μM and a cytotoxic concentration (CC50) greater than 100 μM [6].
Chloroquine and quinine are antimalarial drugs derived from quinoline alkaloids [7]. Quinoline (Fig.1a) is a heterocyclic aromatic organic compound having a molecular formula of C9H7N.
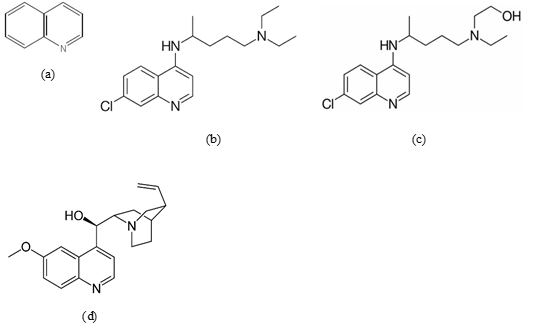
Fig.1 2D structure of (a) quinoline; (b) chloroquine; (c) hydroxychloroquine; and (d) quinine (built using MarvinSketch of ChemAxon)
Chloroquine (Fig. 1b), hydroxychloroquine (Fig. 1c), and quinine (Fig. 1d) possess a similar basic quinoline structure. However, the difference is that chloroquine belongs to the 4-aminoquinoline class, while quinine belongs to the 4-quinolinemethanol structure [7].
Quinine is an alkaloid compound contained in the Cinchona bark [8]. Quinine remained a mainstay of malarial treatment up to the 1920s, until a more effective synthetic anti-malarial drug, chloroquine, was widely used, especially starting in the 1940s [9].
In addition to having structural similarities, chloroquine and quinine also indicate the same mechanism as antimalarial drugs. For its survival, Plasmodium falciparum requires nutrients obtained by digesting hemoglobin in the acidic vacuole. The hemoglobin that is degisted in addition to producing amino acids which are nutrients for parasites also produces a toxic substance called ferriprotoporphyrin (FP-IX). Chloroquine and antimalarials containing other quinoline rings form complexes with FP-IX in vacuoles. The FP-IX drug complex is very toxic and cannot combine to form pigments. The toxin complex drug FP-IX inhibits food intake thus causes the deceases of the parasite due to lack of food [10,11]. The FP-IX chloroquine complex also interferes with the permeability of the parasitic membrane and membrane proton pump. Another mechanism of action is to interconnect with parasite DNA and inhibit DNA polymerase (quinine). Chloroquine is also a weak base so that the entry of chloroquine into an acidic food vacuole increases the pH of the organelle. Changes in pH will inhibit the activity of aspartic protease found in food vacuoles so that parasitic metabolism is disrupted [12].
This work aims to study the binding mode of quinine, chloroquine, and hydroxychloroquine in the peptidase domain of the ACE2 receptor by molecular docking. Therefore, the results of this study could be utilized as an initial reference for the treatment of patients infected with COVID-19.
Materials and Methods
Hardware used was MacBook Pro (13-inch, Mid 2012) embedded with macOS Mojave 10.14.6, 2.5 GHz Intel Core i5 processor, a memory of 16 GB 1600 MHz DDR3, and Intel HD Graphics 4000 1536 MB. Softwares used were MarvinSketch of ChemAxon 17.11.0 (Academic License), LigandScout 4.1.4 (Universitas Padjadjaran License), AutoDock Tools 1.5.6 and AutoDock Vina 1.1 (Freeware).
Protein preparation
Protein target was the crystal structure of human Angiotensin Converting Enzyme (ACE2) receptor (PDB code: 6VW1; resolution 2.68 Å; R-value free 0.229, R-value work 0.197), downloaded from https://www.rcsb.org/structure/6VW1 [13]. LigandScout was employed to automatically activate the PDB interpretation algorithm and showed the complex in the macromolecule view. Its ligand-protein interaction was studied.
Ligand preparation
The 2D structure of all test ligands, e.g. chloroquine, hydroxychloroquine, and quinine, were built using MarvinSketch, then were converted to a 3D structure using LigandScout. Geometry optimization was performed using MMFF94, which is intended to produce accurate geometric structures [14].
Molecular docking
Molecular docking was carried out for all ligands in the peptidase domain of ACE2 receptor. AutoDockVina was used for molecular docking which is already embedded in the LigandScout software [15,16].
Results and Discussion
The coordinate of ACE2 receptor LBD are x = 85, y = -10, z = 180 (grid box dimension: x=75 points, y=75 points, z=75 points, spacing: 0.375 Å) which proves the presence of Asn33A, Glu37A, Asp38A, Leu39A, Tyr41A, Gln42A, Lys353A, Arg357A.
The docking of all test compounds revealed that chloroquine, hydroxychloroquine, and quinine can interact with important amino acid residues, e.g. Lys353, in the peptidase domain of the ACE2 receptor. Of the three compounds, quinine shows the strongest affinity to the ACE2 receptor (-4.89 kcal/mol) followed by hydroxychloroquine (-3.87 kcal/mol), and chloroquine (-3.17 kcal/mol), respectively (Table 1 and Fig.2).
|
Table 1. The binding affinity of quinine, chloroquine, and hydroxychloroquine with ACE2 receptor |
||||
|
No. |
Compound |
Binding Affinity (kcal/mol) |
Amino Acid Residues Interaction |
Type of Bonding |
|
1. |
Chloroquine |
-3.17 |
His34 |
Positive Ionizable |
|
|
|
|
Glu37 |
Hydrogen Bond, Positive Ionizable |
|
|
|
|
Lys353 |
Hydrogen Bond |
|
2. |
Hydroxychloroquine |
-3.87 |
Asn33 |
Positive Ionizable |
|
|
|
|
His34 |
Hydrogen Bond, Positive Ionizable |
|
|
|
|
Lys353 |
Hydrogen Bond |
|
3. |
Quinine |
-4.89 |
His34 |
Positive Ionizable |
|
|
|
|
Glu37 |
Positive Ionizable |
|
|
|
|
Lys353 |
Hydrogen Bond |
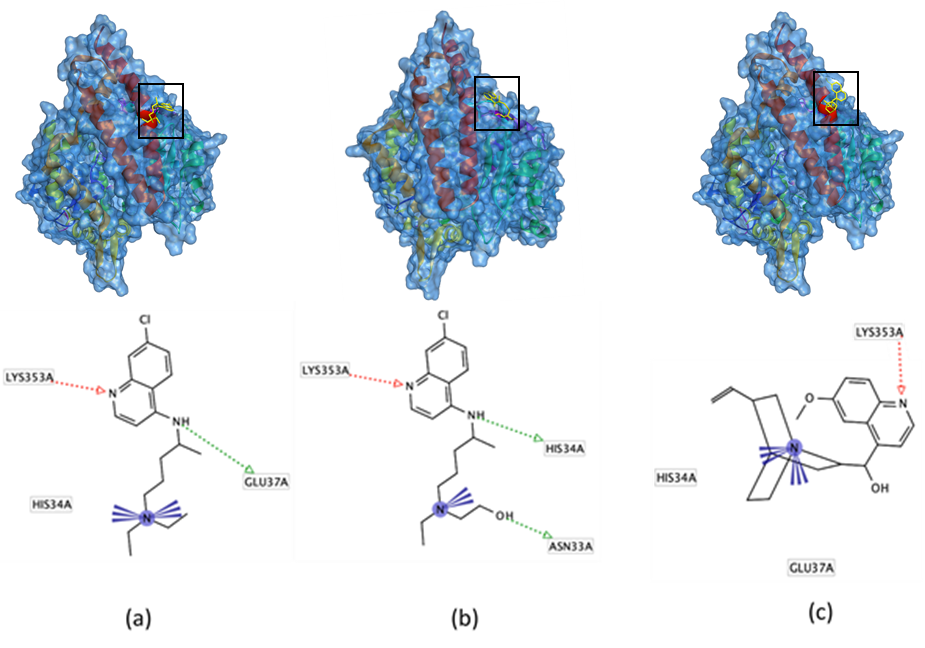
Fig.2: Docking pose and interaction of chloroquine (a), hydroxychloroquine (b), quinine (c) with amino acid residues in the peptidase domain of ACE2 receptor. The hydrogen bond is shown as a dotted-arrow (HBA: red; HBD: green), while the ionic bond is visualized by violet color.
Four ACE2 receptor residues (residues 31, 35, 38, and 353) are considered important in the infection of SARS-CoV because these residues are the site where the virus binds [17]. Our molecular modeling demonstrated that chloroquine, hydroxychloroquine, and quinine interact with residue Lys353 in the peptidase domain of the ACE2 receptor, thus preventing the binding of the virus to human ACE2 receptor. This result corresponds with that of a recent study by Wang and coworkers (2020) which reported that chloroquine could inhibit COVID-19 infection [18]. Chloroquine was also reported could inhibit the SARS-CoV-2 binding to target cells [4].
Conclusion
Chloroquine, hydroxychloroquine, and quinine interact with the important residue Lys353 in the peptidase domain of ACE2 receptor, furthermore, chloroquine and hydroxychloroquine were proven could block the infection of the SARS-CoV-2 virus. Of the three drugs, quinine shows the strongest affinity to the ACE2 receptor (-4.89 kcal/mol), thus potential to be used as COVID-19 antidote. This study will add more insights to the mechanism of quinoline-based antimalarial drugs in inhibiting the infection of the SARS-CoV-2 virus.
Conflicts of Interest
The authors declare that they have no conflict of interests.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
References
- Hoffmann, M., Kleine-Weber, H., Krüger, N., Müller, M., Drosten, C., Pöhlmann, S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv, 2020; 1–5.
- Wrapp, D., Wang, N., Corbett, K.S., Goldsmith, J.A., Hsieh, C.-L., Abiona, O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, 2020; 367(6483), 1260–1263.
- Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., Zhou, Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.), 2020; 367(6485), 1444–1448.
- Devaux, C.A., Rolain, J.-M., Colson, P., Raoult, D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19?. International Journal of Antimicrobial Agents, 2020; 1–6.
- Gao, J., Tian, Z., Yang, X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. BioScience Trends, 2020; 14(1), 72–73.
- Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 2020; 30(3), 269–271.
- Nqoro, X., Tobeka, N., Aderibigbe, B.A. Quinoline-Based Hybrid Compounds with Antimalarial Activity. Molecules (Basel, Switzerland), MDPI: 2017; 22(12), 1–22.
- Gachelin, G., Garner, P., Ferroni, E., Tröhler, U., Chalmers, I. Evaluating Cinchona bark and quinine for treating and preventing malaria. Journal of the Royal Society of Medicine, 2017; 110(1), 31–40.
- Winstanley, P. Handbook of drugs for tropical parasitic infections (2nd edition). Transactions of the Royal Society of Tropical Medicine and Hygiene, 1996;
- Fitch, C.D. Chloroquine resistance in malaria: a deficiency of chloroquine binding. Proceedings of the National Academy of Sciences of the United States of America, 1969; 64(4), 1181–1187.
- Krogstad, D.J., Gluzman, I.Y., Kyle, D.E., Oduola, A.M.J., Martin, S.K., Milhous, W.K., et al. Efflux of chloroquine from Plasmodium falciparum: Mechanism of chloroquine resistance. Science, 1987; 238(4831), 1283–1285.
- Croft, S.L. Principles of pharmacology: A tropical approach; Cambridge University Press: 1992;
- Shang J, Ye G, Shi K, Wan YS, Aihara H, L.F. Structure of 2019-nCoV chimeric receptor-binding domain complexed with its receptor human ACE2. 2020;
- Halgren, T.A. MMFF VI. MMFF94s option for energy minimization studies. Journal of Computational Chemistry, 1999; 20(7), 720–729.
- Trott, O., Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 2009; 31(2), 455–461.
- Wolber, G., Langer, T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. Journal of Chemical Information and Modeling, 2005; 45(1), 160–169.
- Li, F. Structural Analysis of Major Species Barriers between Humans and Palm Civets for Severe Acute Respiratory Syndrome Coronavirus Infections. Journal of Virology, 2008; 82(14), 6984–6991.
- Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro; 2020; 269–271
Contact SPER Publications
SPER Publications and
Solutions Pvt. Ltd.
HD - 236,
Near The Shri Ram Millenium School,
Sector 135,
Noida-Greater Noida Expressway,
Noida-201301 [Delhi-NCR] India
